... description
Introduction
April 16, 2010
We consider sparse and identifiable linear latent variable (factor) and linear Bayesian network models for parsimonious analysis of multivariate data. We propose a computationally efficient method for joint parameter and model inference, and model comparison. It consists of a fully Bayesian hierarchy for sparse models using slab and spike priors (two-component δ and continuous mixtures), non-Gaussian latent factors and a stochastic search over the ordering of the variables. The framework which we call SLIM (Sparse Linear Identifiable Multivariate modeling) is validated and bench marked on artificial and real biological data sets. SLIM is closest in spirit to LiNGAM, but differ substantially in inference, Bayesian network structure learning and model comparison. In comparisons SLIM performs equally well or better than LiNGAM with comparable computational complexity. We attribute this mainly to the stochastic search strategy used, and to parsimony (sparsity and identifiability) which is explicit part of the model. We propose two extensions to the basic iid linear framework: non-linear dependence on observed variables called SNIM (Sparse Non-linear Identifiable Multivariate modeling) and allowing for correlations between latent variables called CSLIM (Correlated SLIM) for the temporal and/or spatial data.
SLIM in a nutshell
April 16, 2010
Starting from a training-test set partition of data {X,X★}, our framework produces factor models CL and DAG candidates B with and without latent variables CL that can be compared in terms of to how well they fit the data using test likelihoods L. The variable ordering P needed by the DAG is obtained as a byproduct of a factor model inference. Besides, changing the latent variables Z produces two variants of SLIM.
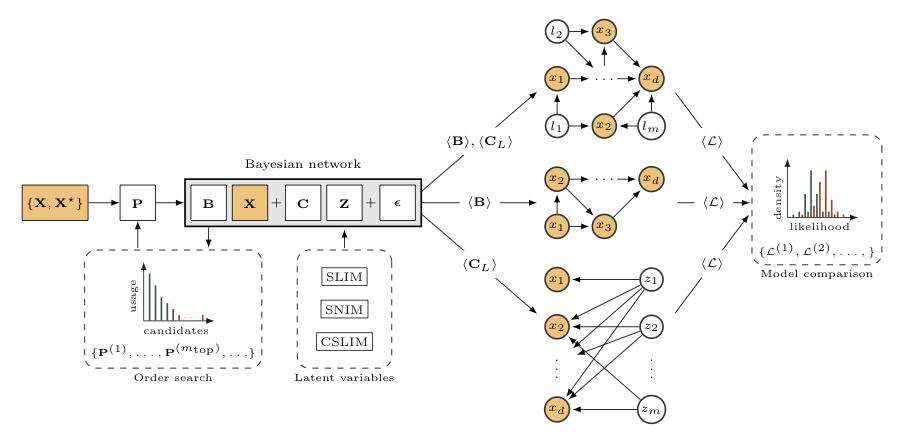
Highlights:
- Sparse factor models including over-complete representations
- Pure DAGs
- DAGs with latent variables
- Non-linear DAGs
- Support for correlated data (time series, spatial data)
- Inference with missing data (not fully coded: non-iid data only)
- Model comparison using predictive densities
... software
Matlab package
April 16, 2010
Set of functions and demo scripts implementing all variants of SLIM including its non-linear version, SNIM. The functions are fairly optimized for speed, not for storage and unfortunately the code is scarcely documented, however, it must be easy to follow since all the required conditional posteriors appear in the appendix of the paper.
zip
C package
April 16, 2010
Set of functions, terminal application and Matlab interface functions implementing all variants of SLIM including its non-linear version, SNIM. The code is single-threaded and optimized for speed. All posterior samples are dumped to a file so memory requirements are greatly reduced. The code uses GSL so must be fairly easy to follow. We tested the code on Mac and Linux machines both 32 and 64 bits using gcc without any inconveniences.
soon
... references